In the development of an active pharmaceutical ingredient (API), it is important to design a formulation that provides adequate stability over long term storage with recommended handling conditions. A liquid formulation is an easy and economical way to handle the product during manufacture and is convenient for the end user. Many proteins prove difficult to formulate as a stable solution and often require lyophilisation. Recombumin, a recombinant human albumin, has been developed to aid in the development of liquid formulations for unstable API's.
Recombumin is produced by Novozymes Biopharma UK Ltd, and is a Saccharomyces cerevisiae yeast- derived protein. It is structurally identical to Human Serum albumin (HSA). Recombumin itself is formulated at pH7 and has been shown to have a shelf life of greater than 5 years at 5°C [1].
This is one of several application notes that use Recombumin or Albucult (both samples are recombinant HSAs but as different formulations) from Novozymes Biopharma UK to explore how light scattering techniques can be used to evaluate formulation stability [2, 3, 4]. This application note studies the thermal stability of Recombumin over the range of pH conditions which API's may be formulated, by following its unfolding and aggregation with increasing temperature. This knowledge allows determination of the most appropriate pH range for this molecule to provide a stable liquid formulation.
Recombumin samples at different pH values were kindly provided by Novozymes Biopharma UK. The Recombumin samples provided were diluted from the 20% stock solution 20 fold to 10mg/ml in the following 25mM ionic strength buffers; citrate pH 3, 4, 5, and 6, phosphate pH 6, 7, and 8, and borate pH 8, 9, and 10. The ionic strength of each sample was adjusted with sodium chloride to be approximately equivalent to 0.9% w/v sodium chloride. The samples were sterile filtered in 2mL aliquots and stored at 5°C for about 6 months prior to the time of measurement.
The samples were measured with the Zetasizer APS - an automated plate sampler system which allows the unattended dynamic light scattering (DLS) measurements of multiple samples.
All samples were filtered with a 0.02µm filter before measurement to ensure that all larger aggregates were removed before measurement. This is a precautionary step to ensure that all samples have similar low content of larger aggregates. As seen in the application note studying the protein at high concentration, this protein formulation is monodisperse [3]. The removal of larger aggregates and possible other contaminants such as dust particles by filtration is done to ensure that they do not affect the aggregation process of the sample. Aggregation is often a nucleus dependent process, where the presence of aggregates affects the aggregation rate of the sample.
The thermal trend measurements started at 25°C and increased to 85°C, in 1°C steps with an equilibration time of 20 seconds. One DLS measurement was performed at each temperature.
The temperature where the sample starts to aggregate is either called the aggregation temperature, Tagg, or melting temperature, Tm. It is important to note, this is not the same melting point as measured in, for example, circular dichroism measurements which reports on the loss of secondary structure elements [4].
The change in the protein monomer hydrodynamic radius over the temperature range measured is shown in figure 1 for the Recombumin samples formulated at pH 3 to 10. It is clear that most of the samples show similar Tm of 67 to 68°C. The two samples formulated at pH 6 (citrate and phosphate buffer) show the greatest thermal stability as they do not start to aggregate until above Tm of 74°C.
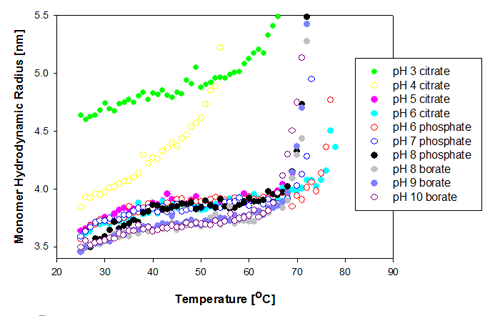
|
The two samples that differ significantly from the rest in their behavior are those prepared at pH 3 and pH 4. The pH 4 sample is the first to unfold, and its unfolding rate differs from the other samples'. The increased sensitivity to temperature indicates this is the least stable of all of the samples. The Recombumin at pH 3 starts with a much larger radius than any of the other samples. This is probably due to a significant amount of dimers present in the sample. This was shown to be the case with SEC data [2]. Dimers and monomers are too close in size for DLS to separate them into two peaks and therefore the peak mean values are larger. Molecular weight can be converted into hydrodynamic size from well established relationships [5]. From such relationships, a Recombumin monomer would have an expected hydrodynamic radius of 3.57nm whereas the dimer would have a Rh of 4.8nm. In Figure 1, it is seen that the measured hydrodynamic radius is close to 4.8nm, and from that it can be concluded that there are a significant proportion of dimers present in the sample. For detection of small oligomers see application note [6].
As the dimer is smaller than the filter pore size used (20nm), it is expected to pass through the filter.
The aggregation temperature in DLS is determined by a linear fit to the scattering signal, to detect where the signal starts to increase due to the increased scattering from aggregated molecules. The aggregation temperatures for the Recombumin samples are shown in figure 2. These data confirm that the size change is directly related to the change in scattering as it is Recombumin in pH 6 that is the most thermally stable sample. Thereafter, the samples of Recombumin at pH 5, 7, 8 and 9 show very similar melting points and hence thermal stability. These data indicate that the Recombumin sample is stable within this pH range.
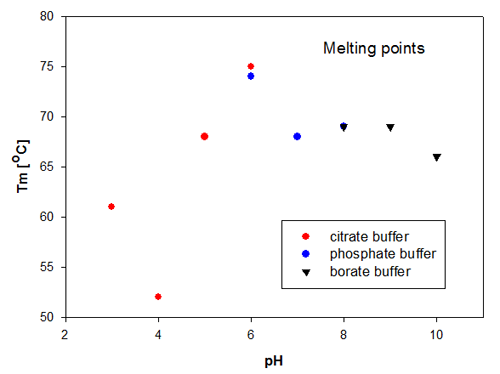
|
The end points of the pH range, i.e. pH10 and pH 3, show lower melting points. The significantly lowest thermal stability is exhibited by Recombumin at pH 4 which starts aggregating at 52°C. It is noticeable that the determinations of melting temperatures/aggregation temperatures are very reproducible in the cases where the Recombumin has been formulated in two different buffers with the same pH, see samples at pH 6 and pH 8.
To illustrate the predictive capabilities of the thermal trend measurements with DLS, the melting point temperatures were compared to long-term stability measurements where the same samples have been measured at 6 months and then 9 months after preparation. The samples were kept at a storage temperature of 2 to 8°C before and in between these measurements. Figures 3a and 3b show the changes in the sample indicated by the hydrodynamic size, where it is clear that the samples at pH 3 and pH 4 are the least stable Recombumin samples in the pH range explored.
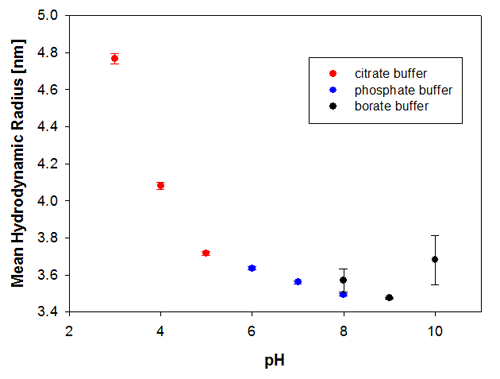
|
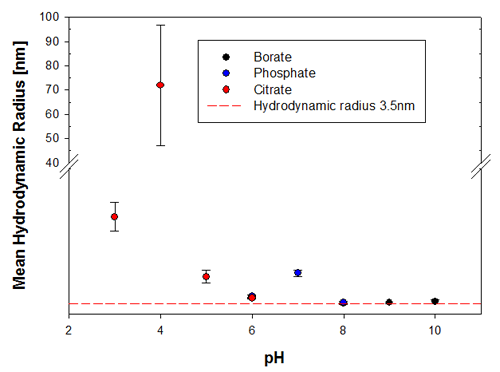
|
As shown in this application note, thermal trends by DLS are useful both for predicting the stability of a protein formulation, as well as for monitoring how the protein responds to thermal stress. This information is useful in the design of production processes and sample handling recommendations.
[1] "Investigating the stability of recombinant human serum albumin under a range of pH conditions with both time and temperature"
Poster presentation by Mr Karl Nicholls, May 2010, BPI Europe 2010, Vienna - Austria
[2] MRK1616 App note GP.HPLC & DLS
[3] MRK1614 App note high conc protein data
[4] L Masino, SR Martin and PM Bayley "Ligand binding and thermodynamic stability of a multidomain protein, calmodulin" in Protein Science (2000) 9: 1519-1529
[5] P Claes, A Kennedy and P Vardy "An on-line dynamic light scattering instrument for macromolecular characterization" in "Laser light scattering in biochemistry" SE Harding, DB Sattelle and VA Bloomfield (eds) pp 66-76 RSC (1992)
[6] MRK1615 app note small oligomer detection