Biopharmaceuticals are drugs made from biological molecules such as proteins. Protein activity is often defined by its molecular weight and aggregation state. Light scattering is more and more commonly being used to measure the molecular weight of proteins, their oligomers and their aggregates. SEC-MALS is one of the more common implementations of light scattering for protein measurements.
Malvern’s Viscotek SEC-MALS 20 is a multi-angle light scattering instrument with 20 detection angles for the measurement of protein absolute molecular weight and oligomeric state. It can also be used to study the molecular weight and size of protein aggregates, as well as for the analysis of synthetic and natural polymers.
The Waters Alliance® system is frequently used in protein separations and is popular throughout the biopharmaceutical environment, particularly due to its powerful software, Waters Empower®. Adding advanced detectors to most SEC systems is usually possible thanks to the generic analog and digital connections that can be used to transfer signals between devices but software packages are often incompatible.
In this application note, we show that Malvern’s OmniSEC software can now communicate with Waters Empower® for the seamless exchange of sample sequences. This allows OmniSEC to follow the injections performed by a Waters SEC system and to synchronize data collection. A selection of proteins was used to demonstrate the measurement of protein molecular weight, oligomeric and aggregation state.
The Viscotek SEC-MALS 20 was connected to the Waters Alliance® system comprising an e2695 pump, degasser and autosampler module and an e2414 RI detector (figure 1). The autosampler trigger signal was connected to synchronize injections and data collection. The RI analog signal out from the RI detector was connected to one of the analog inputs on the SEC-MALS 20. The fluidic connections went from the column, to the SEC-MALS 20 and then to the RI detector. The RI detector was plumbed last in line to protect the flow cell from any increase in back-pressure.
Waters Empower® and Malvern’s OmniSEC software were installed on the same PC. The ‘sample set method’ was created in Empower® and set to run. OmniSEC then loaded the sequence information from Empower ®
The separations were performed on a Malvern PLS5030 SEC column running PBS at 1 mL/min. These columns are silica-based columns for the separation of proteins. Their high resolution and low particle shedding makes them ideal for protein light scattering measurements.
Bovine serum albumin (BSA) was run as a molecular weight standard and used for detector response calibration and normalization. Two test proteins, pepsin and trypsin inhibitor, were dissolved in the mobile phase. The trypsin inhibitor was heated in a microwave for 10 seconds to stimulate severe aggregation. From experience, pepsin samples already contain significant levels of aggregates. The two samples were then injected (100 µL) at concentrations of approximately 2 mg/mL. The results were then analyzed in OmniSEC.

|
The BSA monomer calibration results were within specification and the results were shown to be accurate by correctly measuring the molecular weight of the dimer. The test proteins, pepsin and trypsin inhibitor, were then measured.
Figure 2 shows a chromatogram of pepsin which contains three peaks. The most well defined peak eluting at 9.7 mL is clearly the monomer and its molecular weight has been correctly measured to be 35 kDa. The earlier eluting peak has a very high and polydisperse molecular weight and clearly results from some large aggregates in the sample. The less well defined peak eluting after the monomer has a lower molecular weight and results from some degraded protein. This sample therefore clearly shows that the SEC-MALS 20 detector, connected with the Waters Alliance® system, can measure monomer, aggregate and degradation product molecular weight (figure 2).
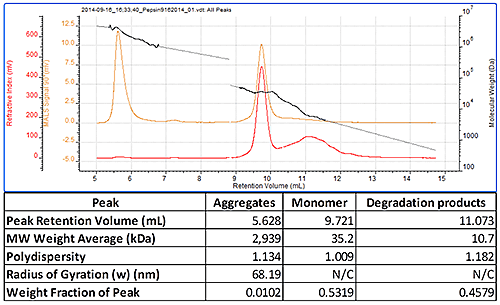
|
Figure 3 shows a chromatogram of trypsin inhibitor, which had been heated in a microwave to induce aggregation before the measurement. This sample showed the monomer peak whose molecular weight was measured to be 27 kDa. There were a number of aggregate peaks which were all analyzed as a single peak. This had very a high molecular weight of approximately 450 kDa and was polydisperse as is typical for such aggregates.
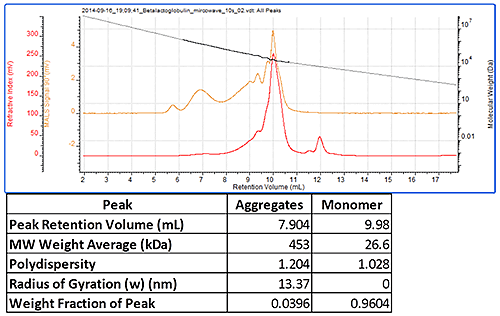
|
MALS measurements can also be used to measure the molecular size (Rg) of protein aggregates if they are over the lower limit of the size measurement, which is approximately 10 nm. This can be measured by studying the angular dependence of the scattered light. Here, the sizes of the pepsin and trypsin inhibitor aggregates were compared. Pepsin’s aggregates had a molecular weight of almost 3 million Da and a very large measured size of 68 nm. On the other hand, trypsin inhibitor’s aggregates were approximately 450 kDa but their size was much smaller at 13 nm. The differences in the angular dependence graphs can clearly be seen in figure 4, which shows a single data slice of the two samples at approximately the same molecular weight (10,000 kDa). Despite the similar molecular weight, the pepsin aggregate has a measured size of 75 nm compared with trypsin inhibitor's 15.5 nm.
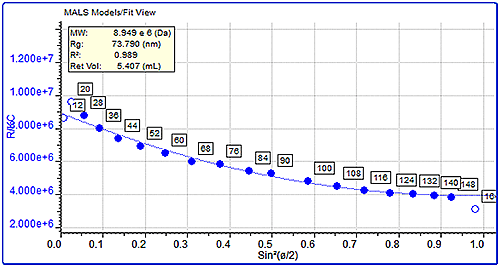
|
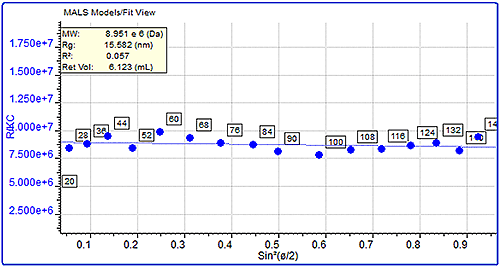
|
Here, we have shown that it is possible to easily and quickly connect the Viscotek SEC-MALS 20 detector (or indeed any other Viscotek detector) with a Waters Alliance® system and to synchronize data collection with Waters Empower® using Malvern’s OmniSEC software.
The molecular weights of two different proteins, pepsin and trypsin inhibitor, were successfully measured using this setup. These measurements demonstrated the range of measurable molecular weights, from the large aggregates in the samples to the small degradation products in the pepsin sample.
It was also possible to measure the sizes of the aggregates within these samples. While the trypsin inhibitor aggregates were relatively small, the pepsin aggregates were much larger suggesting a more open and possibly fibrous structure.
Although a particular set up is shown in this case, these types of measurements can easily be made by connecting Viscotek advanced detectors to any Waters or any other manufacturers’ SEC system.